[From Deep-Sea Life 11, available here]
Chong Chen (1*), Dilworth Y. Parkinson (2), Julia D. Sigwart (3, 4)
(1) Japan Agency for Marine-Earth Science and Technology (JAMSTEC); (2) Advanced Light Source, Lawrence Berkeley
National Laboratory; (3) Marine Laboratory, Queen’s University Belfast; (4) Museum of Paleontology, University of
California, Berkeley
*E-mail: cchen@jamstec.go.jp
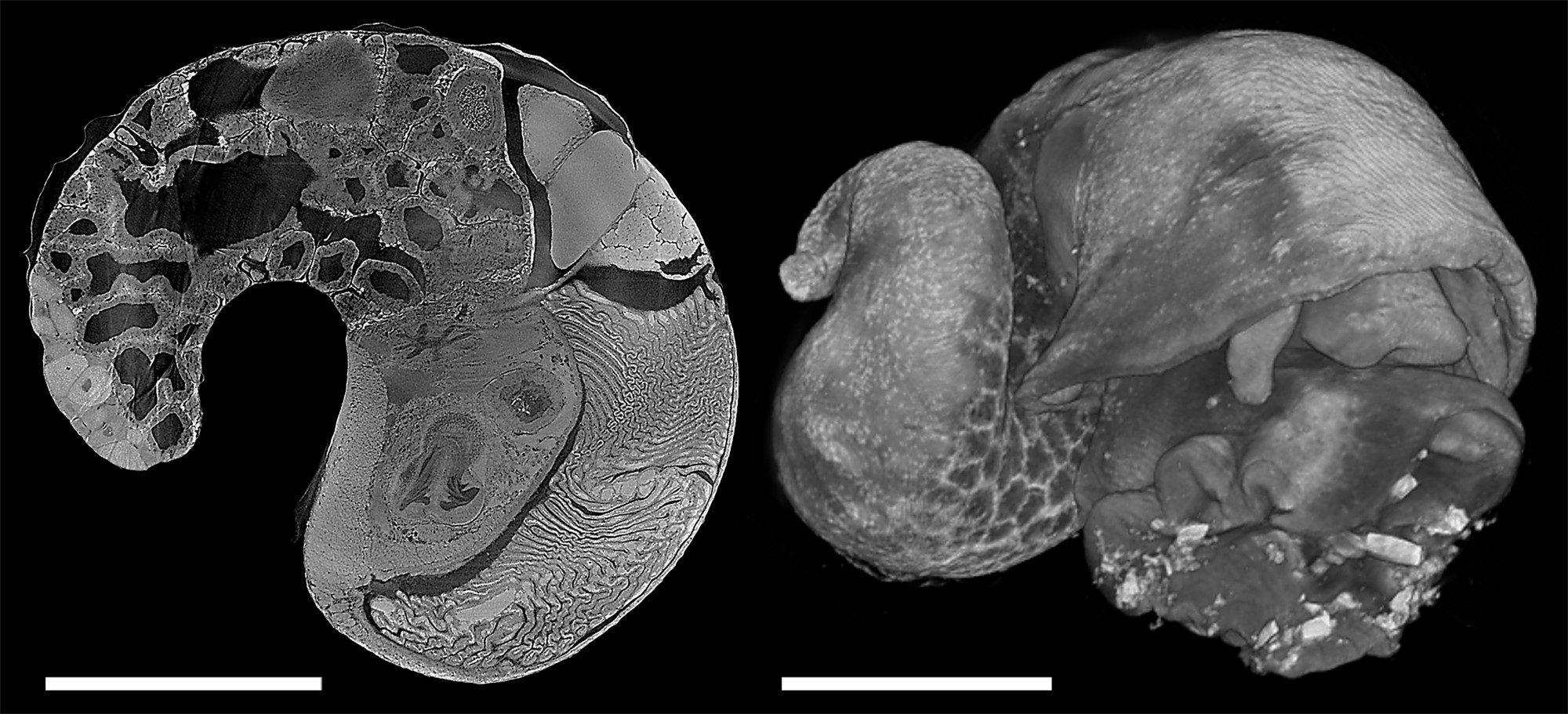
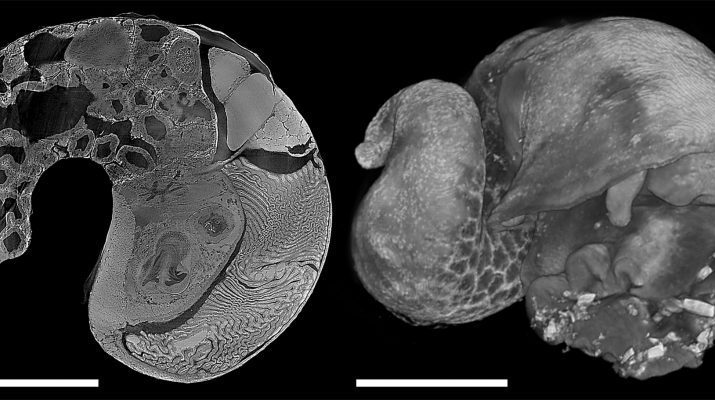
Virtual dissection from 3D digital models use modern technology to provide a window that reveals the way animals are put together. One way to achieve this is digitisation of serial histological sections taken with a microtome, which gives cell-level detail but is labour intensive and destroys the specimen. Commercial micro-CT scanners provides a non-destructive and accurate method for generating 3D data of hard parts such as molluscan shells or foraminifera tests, but the relatively low and unstable x-ray illumination makes it difficult to use for soft parts due to shrinkage and movement associated with long scan times. Hard x-rays from synchrotron light sources, however, have much higher flux which yield high-resolution, low-noise, fast scans. The authors have recently scanned deep-sea molluscs at beamline 8.3.2 at the Advanced Light Source, Lawrence Berkeley National Laboratory in California, USA with wonderful results (Fig. 1). The specimens were fixed in formalin or paraformaldehyde, decalcified, and post-stained with 1% iodine solution prior to the scans; the scans were done at the energy of 23 keV and the collection of 2049 images across 180° rotation took about 20 minutes per specimen. High-resolution data in morphology is vital in understanding the adaptation, autecology, and evolution in complex metazoans which cannot be simply interpreted from omics data, especially for highly derived deep-sea taxa. Synchrotron CT is capable of rapidly delivering exactly that, and with this method it becomes possible to obtain high-quality quantitative anatomical data across many taxa as well as across ontogeny within each taxa towards a comprehensive understanding of adaptive evolution in functional morphology.