As I exit the plane on a hot sticky Los Angeles morning it feels like I’m on a different planet from the ones the cold water corals I study exist in. Having spent the last several years collecting a few thousand corals from museums, research collections and on expeditions I’m finally in a position to start answering some of the large scale questions I have about evolution of corals in the deep sea. Specifically, when have corals around Antarctica diversified? …. Was it in the opening of the Drake passage 40MYA? Or the switch from ‘greenhouse’ to ‘icehouse’ conditions etc? Pinpointing timings with accuracy and understanding the climate history of said times is crucial to teasing apart the drivers of species diversification.
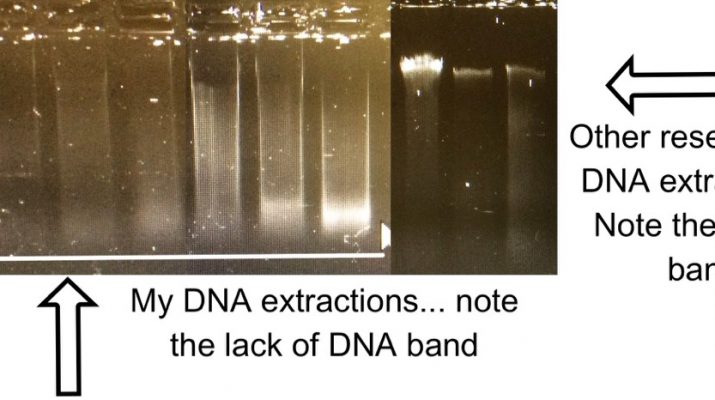
Unfortunately these corals have been collected in such an array of ways, and from many decades ago in some cases, that their DNA quality is, how do I put this, not ideal… The tight high quality band of DNA commonly seen in molecular labs across the world are a rare thing on my gels (see below). If I’m lucky there is a modest smear of DNA in existence. This degraded DNA makes many of the new next generation sequencing techniques an impossibility – UCEs to the rescue (Faircloth et al., 2012)! Ultra-conserved elements are, drum roll, conserved, ultra-conserved some would say. This means that one probe/ marker will be found across a range of animal life; for instance Faircloth et al. used one probe set across amniotes (this covers frogs, crocodiles, chicken and humans…). These conserved regions are flanked by variable genomic areas – this is the elusive variability that I’m after; variability that is a rarity in corals. And, as the DNA needs to be ~800bp in length for this protocol, top quality crème de la crème DNA is not a necessity.
However, the UCE protocol is a protracted one; one with nuance. For this reason I was lucky enough to be welcomed into the famed Cathy McFadden laboratory at Harvey Mudd College and be taught UCE techniques by the indomitable Andrea Quattrini.
Highlights include: visiting the Faircloth lab– incredible collection of bird specimens (see above)! Discovering that there can be “bad” bead mixes (see pictures for proof – bad beads are lighter coloured and pick up far less DNA than darker “good” bead mixes). Chatting with two coral legends about coral. And generally geeking out over the UCE technique. Bring on the DNA extractions! Now I just have to replicate the protocol in the UK… 🙂
References
Faircloth, B.C., McCormack, J.E., Crawford, N.G., Harvey, M.G., Brumfield, R.T. and Glenn, T.C., 2012. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Systematic biology, 61(5), pp.717-726.